I’ve been trying to look into what makes very big cells as laggy as they are. One thing that I came across was organelle splitting, which can sometimes cause noticeable lag spikes as you know. Here’s an issue about it. However, from my testing it seems that those lag spikes are caused by membrane updates instead of the organelle splitting itself. Every organelle split marks the membrane as “dirty”, which leads to the membrane being updated shortly after. The membrane update can take a lot longer than the organelle split itself. Here’s some data I gathered playing through Godot on a non-gaming laptop:
Data
SplitOrganelle takes 0.5-15ms. The time taken was pretty random but generally scaled with size.
Membrane updates:
-
Primum thrivium : 17ms
-
random enemy cell in free build: 70ms (small lag spike)
-
eukaryote with 20 organelles: 120ms (a bit bigger lag spike)
-
big cell with 65 organelles: 450ms (big lag spike)
-
from cache: 0.1ms
Benchmarks
Benchmark results for MicrobeBenchmark v1
Stationary microbes score: 5.255
AI microbes score: 8.627
Spawns until no 60 FPS: 43
Microbe stress average FPS: 54.665
Microbe stress min FPS: 39
Alive microbes: 43
Waiting for microbes to die: 48.74
Microbe deaths minimum FPS: 36
Remaining microbes: 34
Total test duration: 138.2
CPU: Intel(R) Core™ i5-8250U CPU @ 1.60GHz (used tasks: 4)
GPU: NVIDIA GeForce MX150/PCIe/SSE2
OS: Windows
How I collected the data
I couldn’t get normal profiling to work (I’m on Windows and use Visual Studio) so I did the following instead: I measured game ticks before and after the wanted method and then printed the difference to the console. I then converted the ticks (microseconds) to milliseconds. I measured the ticks with Time.GetTicksUsec().
var time1 = Time.GetTicksUsec();
MethodYouWantToMeasure();
var time2 = Time.GetTicksUsec();
GD.Print(time2 - time1);
So what are some takeaways from the data? While SplitOrganelle can take some time, it doesn’t cause the lag spikes. The lag spikes are caused by membrane updates. The more organelles a cell has, the worse the lag spikes are. Membrane updates scaled consistently with organelle count. Updating to a cached membrane is really fast luckily.
So if the membrane update is the culprit, is there something that can be done about it? As it turns out, yes!
I did some tinkering and made my own algorithm that updates the membrane in under a millisecond for small cells and a few milliseconds for big cells. That is up to about 100 times faster. It completely eliminated growth related lag spikes with the aforementioned cells for me. To get lag spikes from organelle splitting now, I’d have to make a cell with at least hundreds of organelles. At that gargantuan scale, it seems that SplitOrganelle becomes the new bottleneck.
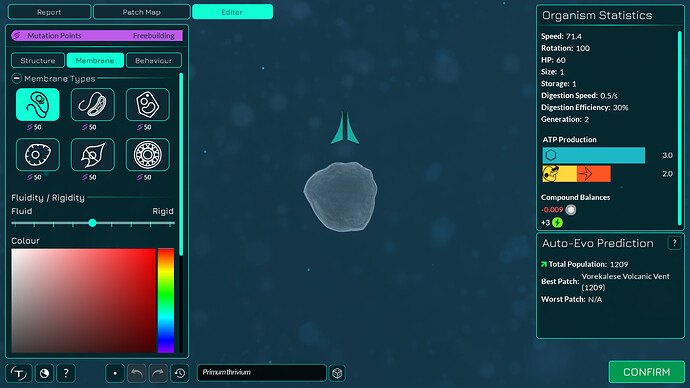
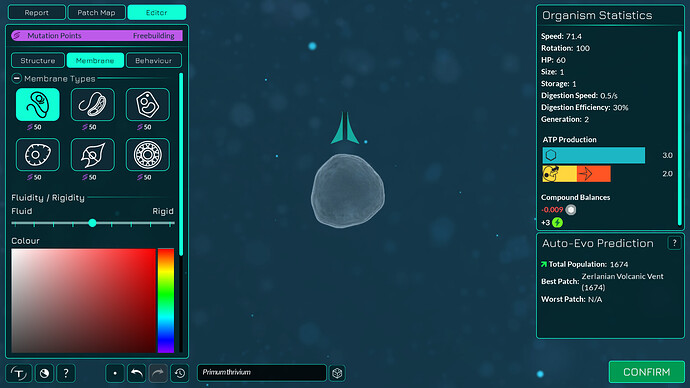
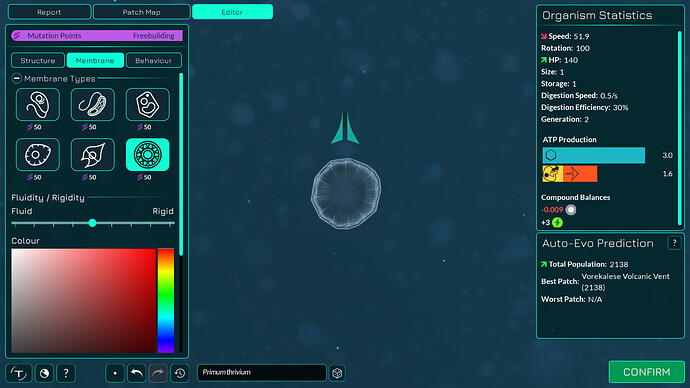
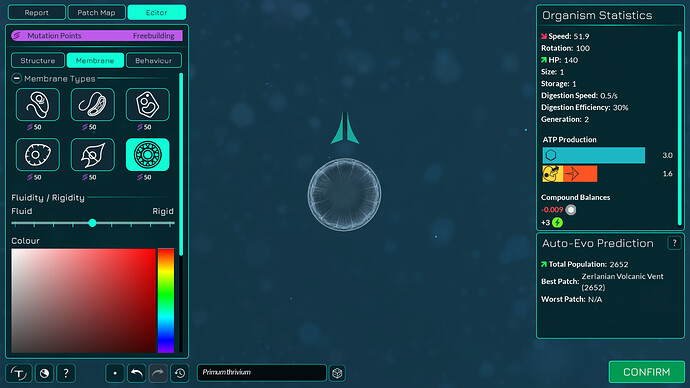
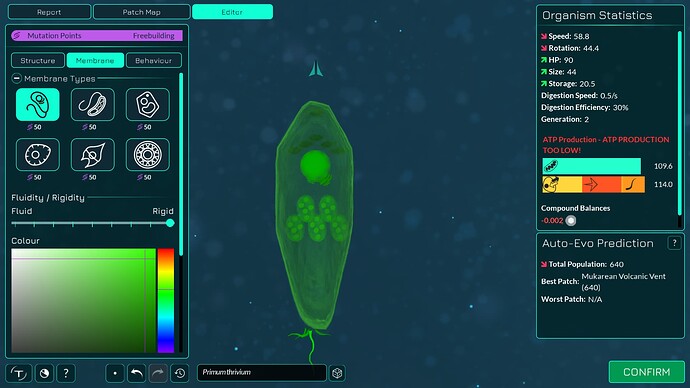
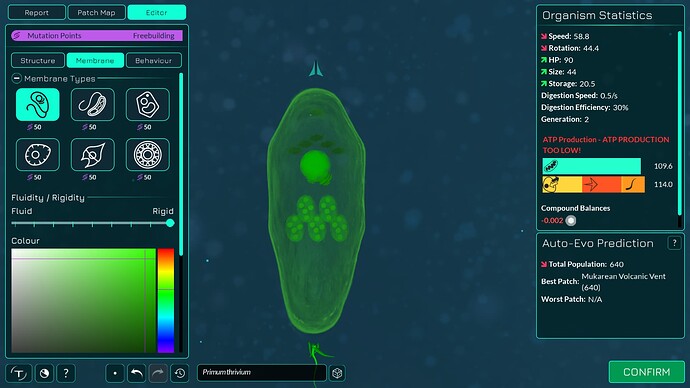
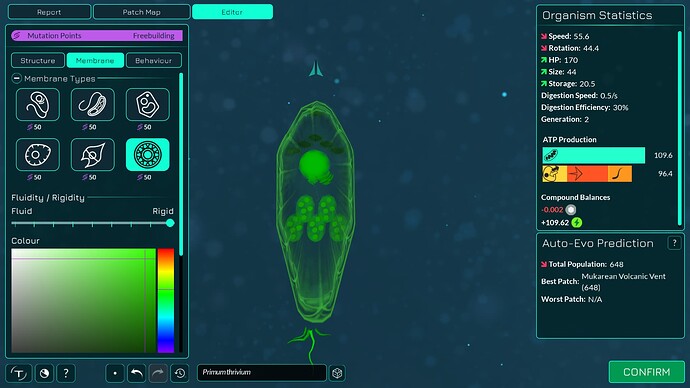
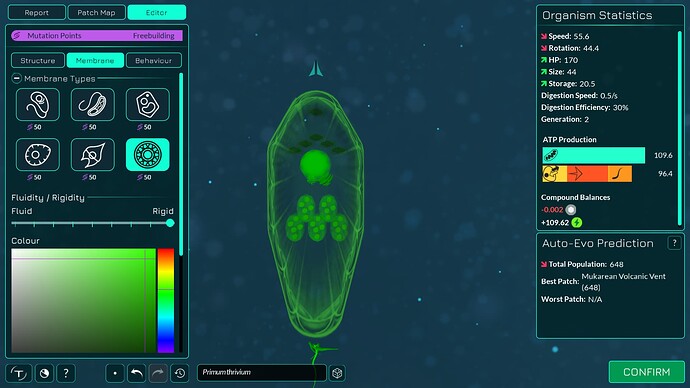
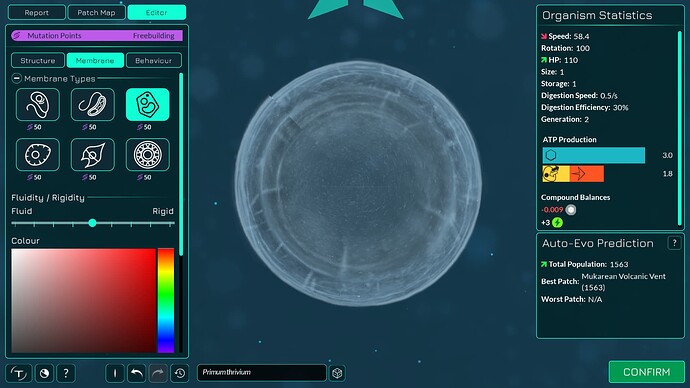
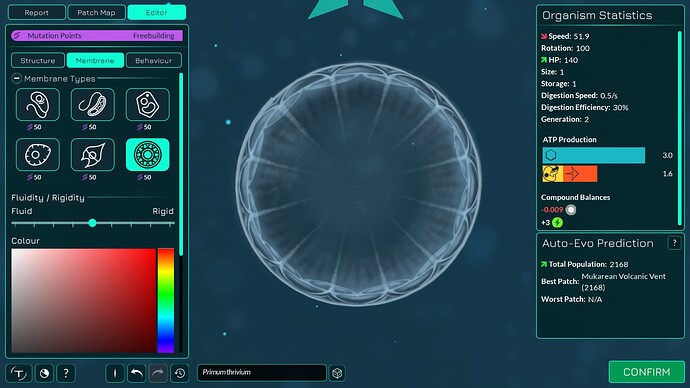
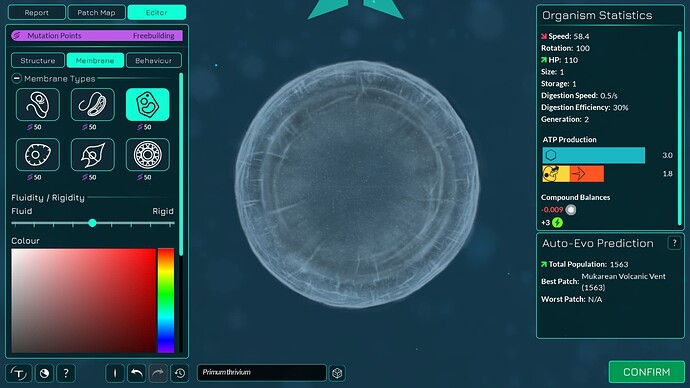
The algorithm works a bit differently from the old one, so it also makes slightly different looking membranes. I tried to keep the end result similar enough. The membranes have more points in them making them smoother. The basic membranes have some waviness to them, the cell walls don’t. Compare these pictures:
The algorithm can also better wrap around organelles, mostly fixing issue 4117. It does that by giving the membrane more distance from organelles if the cell is a eukaryote, which you can kind of see in the “after” pictures. That worked overall better for some reason than giving the membrane hex positions instead of organelle positions.
The algorithm also fixes issue 1274 by making the starting square much larger.
Fixed issue 1274
Before (the cell is a line of 100 Cytoplasm, just invisible because the membrane generated wrong)
After (same cell, no problem)
This is kind of unrelated, but I also noticed that there’s a visible seam in cell walls. I think you know about that, but I couldn’t find a Github issue about it. It can actually be fixed by changing one line of code. If that line is removed, the seam disappears, but that also makes the cell wall textures denser.
Seam
Before (look closely to see the seam in the upper left corner)
Before (notice normal texture density)
After (notice no seam)
After (notice denser texture density)
TL;DR: Membrane updates cause lag spikes. My algorithm gets rid of them. It also fixes a couple other bugs.
I could make a PR for this reworked algorithm if you would like. It would take me a little time to get everything cleaned up for that, and the PR would probably require testing from others to make sure the membrane looks good on a variety of cells. A few questions first:
- Should I make the PR?
- Do the membranes look good?
- If they don’t, what should I change?
- Should I add the cell wall seam fix in?